Open Source Biopharmaceutical Innovation – A Mode of Entry for Firms in Emerging Markets
The open source model provides a valuable framework for collective knowledge production and dissemination. Mirroring the efforts of the open source community that developed Linux, open biopharmaceutical initiatives are enabling companies to access knowledge-based resources critical to drug development. The objective of these initiatives is to preserve the downstream technological opportunities for multiple firms.
As economies in emerging markets enter the biopharmaceutical arena, it is essential that developed economies share not only technological expertise, but also their experiences regarding knowledge production and dissemination. The goals should be to assist these economies to participate on a level playing field with respect to market entry and product development, to protect local knowledge, and ensure fair access to global knowledge as well as technology. Maintaining and building the public domain with particular attention to knowledge that is of benefit to these economies can allow researchers to quickly and cost-effectively access knowledge.
In this paper, two models are developed to understand how open source strategic alliances and open licensing can be used as modes of entry into the biopharmaceutical industry by firms in emerging markets. Case examples and qualitative data are both used to provide a basis for these models.
Introduction
Chesbrough (2003; 2007) explains that inno- vation has become open through the division of labour. In many industries, the vertically integrated organizational structure where innovation is solely an internal activity is gradually being transformed into a more fluid structure integrating internal and external sources of innovation. For example, companies are finding value through the licensing of intellectual property, the development of joint R&D ventures, or other arrangements to exploit technology outside the boundaries of the firm (Chesbrough, 2003; 2007). In the pharmaceutical industry, giants such as Merck and Pfizer have watched as biotechnology upstarts such as Genentech, Amgen, and Genzyme have exploited external discoveries to become major players in this industry. These companies used an open business model in which ideas move from discovery to commercialization through at least two different organizations. (Chesbrough, 2003).
From a knowledge perspective, in the closed model, human capital is employed within the boundaries of the organization. Knowledge is generated within and belongs to the originating firm. The organization’s profit model revolves around the notion that knowledge is discovered, developed, and then embodied within firm-only products (Chesbrough, 2003). Appropriated knowledge is controlled by the originating firm. In the open model, human capital and knowledge are accessed both inside and outside the boundaries of the organization. A firm can profit both from the embodiment of knowledge within internally developed products as well the embodiment of knowledge in products developed by other firms (Chesbrough, 2003).
Open source software development reflects both collaborative production and shared implementation of a technology (Chesbrough et al., 2006). Open source software is considered to be a reaction to the proprietary software model (Lakhani and von Hippel, 2003; Chesbrough et al., 2006). Namely, open source software involves collaborative production and requires free distribution of software source code and the right for others to modify the code. I assert in this paper that open source innovation is a model of open innovation involving collaborative knowledge production and knowledge dissemination with and by participating firms.
Lakhani and von Hippel (2003) discovered in their research three types of incentives driving firm participation in open source software development including: direct utility to the organization from collaborative, open software development e.g., absorptive capacity development and early access to technology; intrinsic benefit from participating in the development of this software e.g., learning a new skill; and signalling one’s abilities in a technological arena to one’s peers or firms. The open source model has provided a valuable framework for collective knowledge production and dissemination beyond the software community. Mirroring the efforts of the open source community that developed Linux, open knowledge networks and other cooperative strategies are enabling biopharmaceutical companies to access disembodied, upstream, knowledge-based resources critical to downstream drug development (Nelson, 1959; Reichman, 2003).
The Human Genome era has emphasized the notion that biological knowledge is complex. Discovery research no longer simply focuses on individual units of knowledge, but considers the behaviour and relationships of all units of knowledge in a particular biological system from a functional perspective (Kitano, 2001; 2002). Genomes are now being described as consisting of complex, intersecting systems rather than unitary collections of separately functioning structures (Hood, 2000; Dutfield, 2003). In this sense, it is possible to observe many similarities to software development. Software is a complex system, developed from many intersecting components (lines of code). Several developers may be required to generate these intersecting lines of code so that the associated processes can emerge and function. Demarcating the lines of ownership in this case can be an onerous task.
As economies from emerging markets enter the biopharmaceutical arena, it is essential that developed economies share not only tech- nology expertise, but also their experiences regarding collaborative knowledge production, technology transfer, and intellectual property management. The goal should be to assist these economies to participate on a level playing field with respect to market entry and product development, to protect local knowledge, and ensure access to global knowledge as well as technology. Researchers and technology transfer officers must therefore, take greater caution in the patenting and licensing of technologies that have significant application in developing and under-developed markets. Maintaining and building the public domain – with particular attention to knowledge that is of benefit to these economies, can allow these researchers to quickly and cost-effectively access knowledge. Open licensing, geographic-based licensing, and assigning fair royalties are additional options being employed to assist researchers in developing economies access technologies that address neglected diseases or local health needs (Chokshi et al., 2006).
As biopharmaceutical knowledge has become increasingly high in complementarity, high in applicability, but low in substitutability, open source innovation, particularly when knowledge exists in disembodied form during the upstream phases of research, can provide multiple firms the opportunity to pursue downstream product development activities. From a mode of entry perspective, open source strategies can further assist firms from emerging markets to enter a technological arena without the onerous upfront costs associated with exporting, developing subsidiaries, pursuing acquisitions or forming joint ventures, as well as encountering transactions costs associated with the sourcing of and contracting for proprietary knowledge (Antonelli, 2003).
I begin by analyzing models of open source innovation from the information technology (IT) sector. Case examples are provided of the use open source IT innovation in emerging markets. I then provide an overview of how the open source model has emerged in the bio-pharmaceutical sector since the completion of the Human Genome Project. Open source based strategic alliances and open licensing are discussed as specific mode of entry options for firms. Case examples and other qualitative data provide the basis for the development of models of these modes of entry for firms in emerging markets.
Open Source Models in the Information Technology Sector
Models of cooperation associated with open standard development and open source software development from the IT sector provide us with valuable insight for cooperative bio-pharmaceutical development. It is important to note that open standard development reflects collaborative technology production between multiple organizations; open source software development entails both collaborative production as well as implementation of a technology.
Open standards are essentially a set of rules for the design of new products. These rules enable coordination between products and components by establishing a common interface to manage their cross-interaction (Chesbrough et al., 2006). Voluntary, non-market standard setting organizations that operate in industries such as software development, where coordination is large, can have a considerable impact on the adoption of a particular technology as an industry standard (Chesbrough et al., 2006).
Open standards create value for consumers by promoting competition between imple- mentations. Firms selling products that implement a standard enjoy less uncertainty associated with the coordination of products (Chesbrough et al., 2006). It is anticipated that firms that produce technologies used to implement a standard, participate in open standard groups to capture the value associated with the development of a new compatibility standard including absorptive capacity development and early access to technology (Cohen and Levinthal, 1990; Chesbrough et al. 2006). Figure 1 is a model of open standard development. The objectives of the open standard setting organization will likely impact the resources and participants that are needed and eventually commit to the development of the open standard. Rules are established to not only manage the technology development process, but also decision making processes, and any associated intellectual property. These rules impact the interactions between the participants and the eventual outcome in terms of standard development.
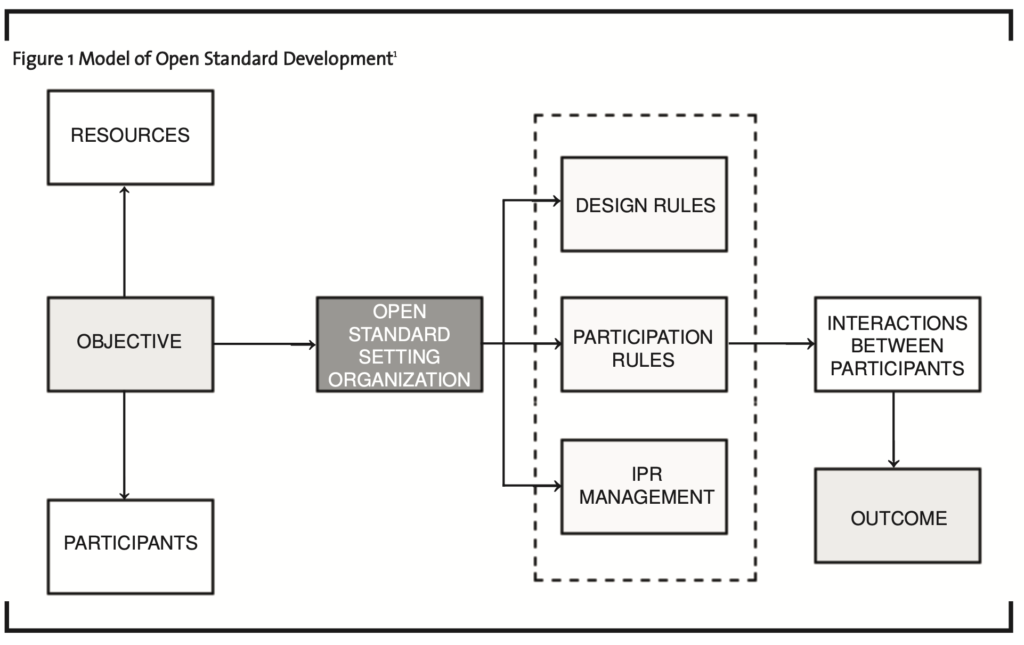
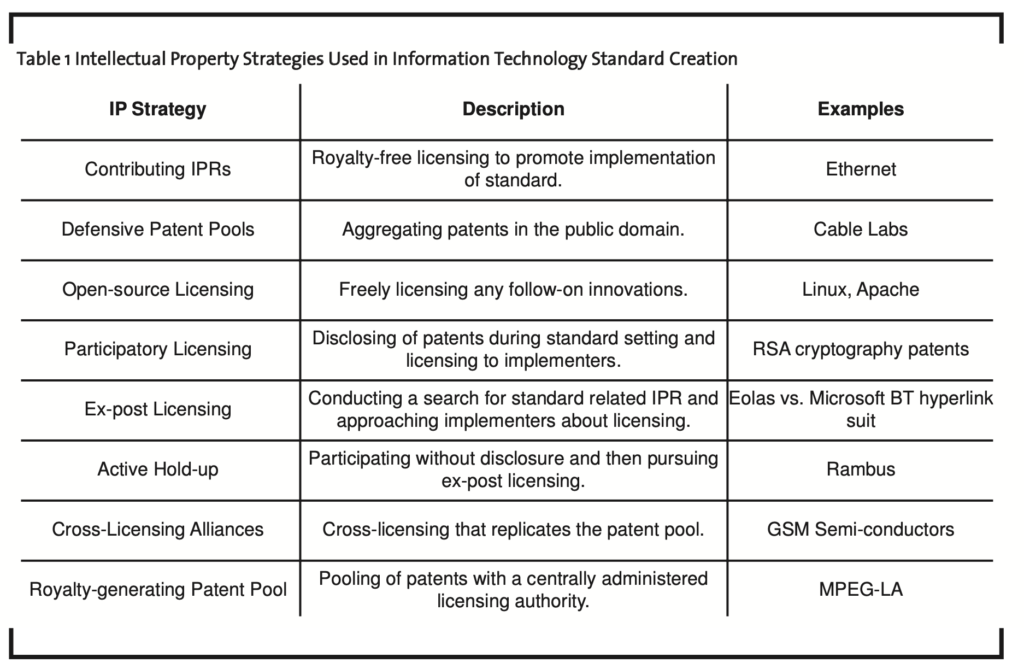
In the management of standard creation, standard setting organizations establish a set of rules and obligations for members as outlined in the charter and bylaws of the organization (Lemley, 2002). Intellectual property rights (IPRs) in open standard development are governed by these rules and address searching for IPRs within member files and or the broader literature, disclosing information within the organization, and licensing of IPR. These rules are essentially designed to prevent members from adopting a standard that entails ex-post hold-ups by patent owners offering a license that likely would not have been accepted ex-ante. Table 1 outlines the intellectual property strategies used in the creation of standards (Chesbrough et al., 2006).
Open source software development reflects both collaborative production and shared implementation of a technology (Chesbrough et al., 2006). Open source software is considered to be a reaction to the proprietary software model, differing from this latter model in terms of intellectual property rights and its production. Namely, open source software involves collaborative production and requires free distribution of software source code and the right for others to modify the code. Two highly visible open source projects are the Linux operating system through the Open Source Development Labs (OSDL) and the Mozilla web browser project. In both cases, firms donate their research to the open source project while exploiting the pooled R&D of the project to enable the sale of related products and services (Chesbrough et al., 2006).
For example, IBM is hoping to take advantage of some of the world’s largest untapped information technology (IT) markets – markets not weighed down by existing proprietary technology – by offering innovation around the adoption of open source solutions such as Linux. IBM has found success pushing open source software into emerging markets because governments in these markets often favour Linux over proprietary technology – finding the idea of proprietary software culturally distasteful (Meredith, 2005).
Linux’ unprecedented growth in the Asia/Pacific region has global ramifications. As alternatives to proprietary systems are adopted and expanded worldwide, the viability of Linux as an operating system (OS) standard continues to increase (Meredith, 2005). In June 2003, IBM jointly established a Linux competency center with the Beijing government in China. IBM and the Beijing government established this center for many reasons, including promoting the usage of Linux by helping organizations port applications to a Linux environment, creating end-to-end Linux solutions, as well as providing training for Linux professionals in China (Meredith, 2005).
A second center in Guangzhou, opened in June 2004, provides software testing, project and technology-management services, as well as professional training courses for local software developers. IBM has instituted similar efforts all over the Asia/Pacific region (Meredith, 2005). According to the Korea Times, in May of 2005, IBM had been in talks with South Korean officials and industrialists about promoting the global open source computer operating system (Meredith, 2005). Similarly driven by cost, licensing issues, and technical issues, a number of companies across India are taking a serious look at the world of free and open source software (Meredith, 2005). IBM officials cite that organizations at all levels find it reassuring to be using an open source system – that is, to see the code powering systems and to understand from the outset the technological issues likely to be encountered with downstream product development.
Sun Microsystems likewise, hopes to use the open source model to help developers use cutting-edge technology to innovate and enab- le the associated countries to move up the worldwide IT value chain. Sun Microsystems provides businesses in emerging economies access to its intellectual property without barriers to adoption, exit, and without barriers of licensing to build their network infrastructure (Sun Microsystems, 2008). Sun Microsystems indicates that governments and educational institutions are warmly embracing open source technologies because countries can move quickly along the IT value chain without the multimillion dollar commitments required to license proprietary technology (Sun Microsystems, 2008).
In February 2008, Sun Microsystems announced its first overseas expansion of its OpenSPARC educational program. The three-year agreement with China’s Ministry of Education (MOE) extends to as many as 10 universities in China this year, and trains 150 teachers each year on Sun’s OpenSPARC designs. As a consequence, Sun Microsystems claims that its business is driven largely by the adoption of open source technologies at the university level and across governments. This broad adoption is thought to be enabling Sun Microsystems to get onto solid ground in a number of emerging markets (Sun Microsystems, 2008).
Research results from several open source studies at UNU-MERIT (United Nations University – Maastricht Economic and Social Research and Training Centre on Innovation and Technology) further suggest that many countries and institutions have made strides in adopting policies to enhance public access to knowledge. In just four years, Extremadura – one of the poorest regions of Spain – successfully invested in creating a free-software society. The model is now being replicated in other poor regions of Spain, as well as in Latin America. In Africa, the University of the Western Cape (UWC) in South Africa has introduced an open learning model spearheaded by the Massachusetts Institute of Technology. The Commonwealth of Massachusetts also successfully introduced OpenDocument – an Open Standard for office applications – which provides important lessons for other regions and countries across the world (Bergstrom, 2006; UNU-MERIT, 2008). Various initiatives at UNU-MERIT hope to use these lessons to assess the effectiveness of several alternative global mechanisms that have been proposed such as Open Source Science and Open Medicine to boost health research and development and broaden access to affordable drugs for the world’s poorest populations respectively (Bergstrom, 2006; UNU-MERIT, 2008).
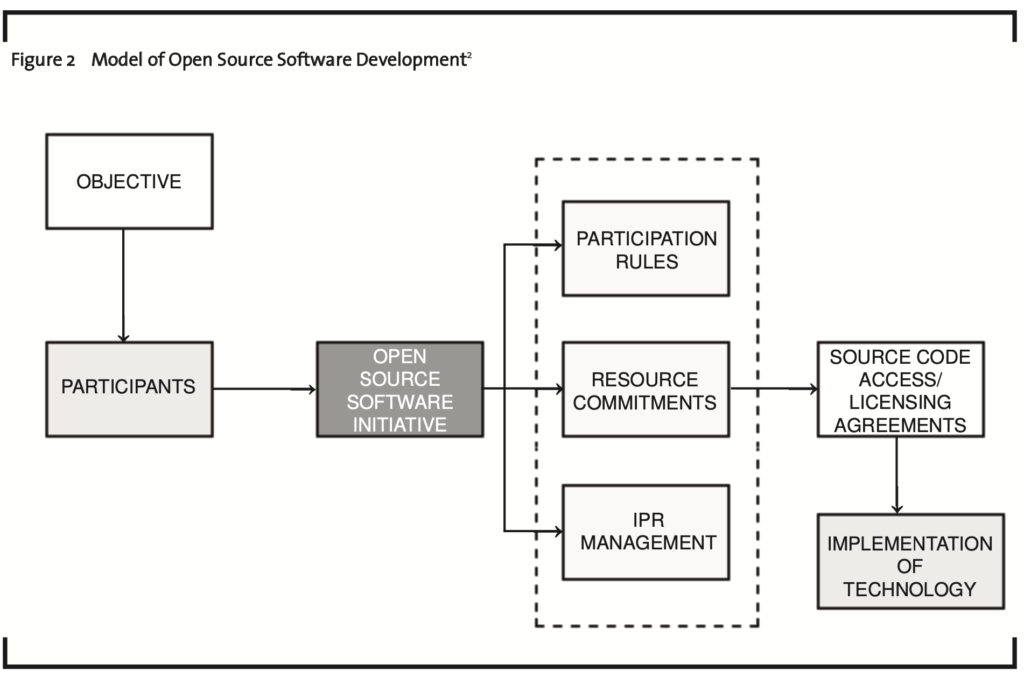
Figure 2 is a model of open source software development. Once again, the objectives of the open source initiative will determine the type of participants that join. In open source software development, the participants are primarily volunteers who are located across different geographic regions using tools to collaborate in source code development. Rules are established once again to not only manage decision making processes and any associated intellectual property, but also resource commitments including technological donations made to the open source initiative. As a result of any donations and development efforts, users are able to use, change, and improve the software, and to redistribute it in modified or unmodified form. Various licensing agreements ensure this open access to source code.
The Emergence of Open Source Innovation in the Biopharmaceutical Sector
The International Human Genome Project catalyzed the open-source movement in genomics-based research. Globally dispersed laboratories jointly collaborated to map and sequence the Human Genome. The resulting data were rapidly deposited into the public domain to ensure an open and level playing field for all researchers. Just as in the case of the previously discussed open standard set- ting organizations and open source software initiatives, leaders of the National Human Genome Research Institute (NHGRI), together with the Wellcome Trust, and academic researchers at the major human genome mapping centres, resolved in February 1996 that all human genomic DNA sequence information generated by centres funded for large-scale human sequencing, should be freely available and in the public domain in order to encourage research and development (Marshall, 1996). NHGRI followed up with an April 1996 policy statement making rapid release of data into public databases a condition for grants for large-scale human genome sequencing (NHGRI, 1996). NHGRI also warned that it would monitor whether grantees were patenting large blocks of primary human genomic DNA sequence and might invoke the exceptional circumstances limitation (to restrict patenting) in future grants (NHGRI, 1996).
A more general statement of Principles and Guidelines for Sharing of Biomedical Research Resources, adopted by the National Institutes of Health (NIH) in December 1999, also attempted to guide NIH grantees in their appropriation activities. The statement outlined that the use of patents and exclusive licenses is not the only, nor in some cases the most appropriate means of implementing the Bayh-Dole Act. Where the subject invention is useful primarily as a research tool, inappropriate licensing practices are likely to thwart rather than promote utilization, commercialization, and public availability (NIH, 1999).
Open source innovation has also flourished in bioinformatics – where software code researchers and databases are traded and pooled on a mutual sharing basis. Researchers indicate that the BioPerl project for example, allowed the development of tools during the Human Genome Project to facilitate the interchange of data amongst laboratories who kept their research in dissimilar formats (Stein, 1996). BioPerl, BioJava, and BioPython — now organized together as the Open Bioinformatics Foundation (OBF), make their work available under standard open source licenses (OBF, 2008). The Bioinformatics Organization further encourages collaborations in bioinformatics development, maintains computational resources, and promotes open access to materials and methods for bioinformatics research and education throughout the world (OBF, 2008).
These efforts in the public sector have similarly encouraged the private sector to promote and participate in open source initiatives. The Single Nucleotide Polymorphisms (SNP) Consortium brought together ten of the world’s largest pharmaceutical firms. Consortium members recognized the SNP map as a pre-competitive, research tool. The Consortium committed to developing a SNP map to assist researchers to identify the multiple genes associated with complex ailments such as cancer, diabetes, vascular disease, and some forms of mental illness. The competitive members viewed the map as a tool to be jointly developed and shared, with open access to the Consortium’s data guaranteed for the public at large. Specifically, the rules established at the outset determined not only knowledge production processes but also included an agreement to relinquish any property rights to the knowledge generated within the Consortium—thereby avoiding any downstream hold-up issues (Davies, 2001).
Then in October of 2004, Novartis, the Broad Institute of MIT, and Harvard announced a joint project to decipher the genetic causes of type 2 diabetes. The collaboration reflected the mission of the Broad Institute to bring together researchers to solve complex problems requiring multi-disciplinary teams and that are difficult to solve in the traditional (isolated) laboratory setting (Lawler, 2004). Companies typically demand that data created in cooperative ventures be kept away from competitors. However, Novartis argued that the benefits of openness would outweigh those of secrecy, and the company placed the genetic variation data it collected on a public web site. While the team did not file patents on the database, it did allow others to patent new therapies or diagnostic tests based on the public information (delaying appropriation to downstream activities) (Lawler, 2004). Novartis’ decision is a signal of an emerging change in attitude toward the appropriation of all forms of biological knowledge —reminding us of reaction that encouraged the development of the open source software model. It is worthwhile to note that in each of the above case examples, organizations are not only benefitting from division of labour typically associated with open innovation (i.e. via collaborative knowledge production), but are also freely accessing knowledge from both inside and outside the boundaries of their own organization (i.e. via adherence to the open source model).
Open Source Biopharmaceutical Innovation as a Mode of Entry
Choosing a mode of entry into a new market and for the purposes of this paper a new technological arena, is a critical decision faced by firms. Firms can choose from a variety of modes, including exports, licensing, wholly owned subsidiaries, acquisitions, and different types of joint ventures. Other modes include subcontracting, associations, and consortia (Malhotra, 2003). In the choice of mode of entry, the knowledge to be transferred is a key issue considered by firms. Namely, protection of knowledge from the threat of opportunism is a primary driver of entry mode choice (Malhotra, 2003). However, it is interesting to observe, that many biotechnology and pharmaceutical companies are promoting and engaging in alliances that are committed to open source drug discovery via cooperative knowledge production and cooperative knowledge dissemination. In the sections that follow, I consi- der both open source based strategic alliances—namely the consortium structure and open licensing as modes of entry into the bio- pharmaceutical arena.
Methodology and Context
The data presented in this paper are sour- ced from a previous study conducted by the author and colleagues. Allarakhia et al. (2008) analyzed 39 open source biopharmaceutical consortia including the likely participants in such initiatives, the rules for participation, the focus of knowledge production activities, and the management of joint knowledge assets. These consortia were visible and significant in their achievements, thereby enabling the researchers to 1) accurately analyze interactions over a reasonable period of time, 2) analyze the policies established with respect to knowledge production and dissemination, and 3) retrieve adequate literature sources for the study. Literature sources analyzed included peer-reviewed journal articles by consortia members or third-party researchers, press releases, consortia websites, publications, and presentations. The researchers also substantiated the data by surveying consortia directors.
It is anticipated that the analysis extended in this paper will allow for an understanding of the structures associated with open source biopharmaceutical initiatives and the development of a model similar to those developed for open standard setting organizations (Figure 1) and open source software initiatives (Figure 2). This model should incorporate how rules and organizational structures encourage entry by firms into such initiatives and in turn the technological arena, how learning is encouraged for participating firms, and how knowledge is disseminated so that firms out- side the open source initiative can pursue pro- duct development opportunities – that is, either at no cost or minimal cost.
In the biopharmaceutical industry, many new drugs hinge upon advances in molecular biology and genetic engineering. As a result, research activity that adheres to the molecular biology paradigm requires network-like alliances between academic institutions, bio-technology companies, and traditional drug manufacturers (Bower and Whittaker, 1992; Powell et al., 1996; Blumenthal et al., 1997). The genomics era has highlighted the need for partnerships that are broad and cross institutional as well as national boundaries. The breadth of upstream research to be conducted to ensure successful drug development, particularly in a decade marked by shrinking pipelines and blockbuster drug patent expirations, has reinforced the need for knowledge-based networks (Reid et al., 2001). Hence, Allarakhia et al. (2008) studied open source based consortia including geographically dispersed participants to understand knowledge production processes in these alliances as well as knowledge dissemination strategies including open licensing employed by consortia members.
Open Source Based Strategic Alliances as a Mode of Entry
As the pharmaceutical industry further transitions into the current post-genome paradigm, the nature of biological knowledge, namely the complementary nature of upstre- am biological knowledge, its complexity in terms of function, and its breadth of application, will encourage the formation of strategic alliances to ensure equitable access to knowledge for future product development. Strong early-mover advantages in drug deve- lopment rest on the ability to rapidly identify, access, and integrate new combinations of knowledge (Antonelli, 2003; Grant and Baden- Fuller, 2004).
Biology knowledge is complex and derives from a variety of scientific and technical disciplines. The molecular level of analysis, the computational nature of discovery research, and the global scale of research, all provide evidence that the drug discovery and development paradigm has changed dramatically. To manage the uncertainties of drug discovery, a new model of cooperation is emerging — the open source consortium (Kitano, 2001; Chokshi et al., 2006). These networks of collaboration are supported by information and communication technologies and are enabling researchers from a variety of disciplines and laboratories to generate and validate biological and chemical knowledge. In these consortia, the issues of data-sharing and intellectual property are closely related. As Chokshi et al. (2006) discuss, consortia must decide in advance what data should be released to the public to ensure equitable downstream access to the data and open opportunities for the development of products; alternatively, in some cases, it may also be necessary to ensure, through the appropriation of data, that downstream incentives for product develop- ment are maintained for consortia members. Rules and policies will determine which option should take precedence in a project and/or consortium. In the sections that follow, I analyze these rules across the 39 selected biopharmaceutical consortia.
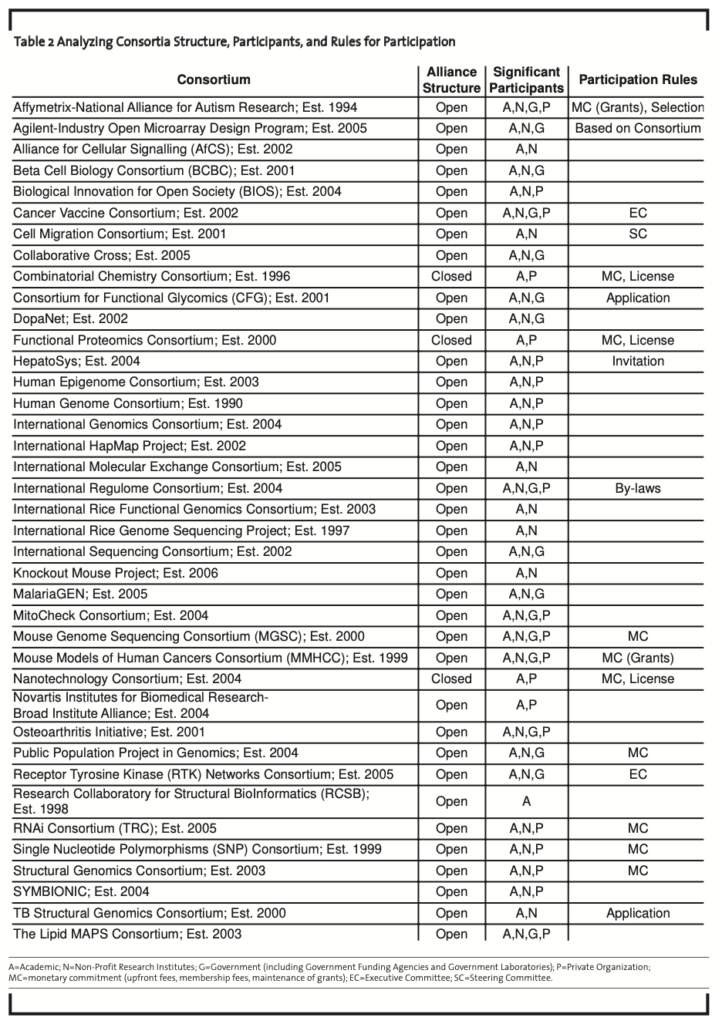
Participant Type. In their analysis, Allarakhia et al. (2008) determined that researchers from academia were present in all 39 consortia; researchers from non-profit research organizations also participated in 34 consortia. In 17 cases, there were government researchers and/or there was government participation via consortium catalyzation or the provision of monetary support. Interestingly in 22 cases, private sector firms were involved to a significant extent (Table 2, see appendix 1). In 6 of these cases, private sector participants were directly or significantly responsible for catalyzing the initiative — namely, the SNP Consortium, the Novartis-Broad Initiative, the Accelrys Combinatorial Chemistry Consortium, the Accelrys Functional Proteomics Consortium, the Accelrys Nanotechnology Consortium, and the Agilent-Industry Open Microarray Design Program (Davies, 2001; Cassier, 2002; Lawler, 2004; Agilent, 2007; Accelrys, 2007). Although 32 out of the 39 consortia were funded by public sources (primarily via government grants), 15 were jointly funded or sponsored by private organizations; and 4, namely the Accelrys Consortia and the Cancer Vaccine Consortium were funded primarily by the private sector participants.
Structure of Open Source Consortia. The decision to participate in open source initia- tives is affected by the degree of accessibility to the associated knowledge. Open access ensures that knowledge is available to all researchers for downstream activities regardless of participation in the initiative. In this case, the possibility of free-riding exists by outside firms who can enjoy the disclosed knowledge at little or no cost (Gintis et al., 2001). Closed access in contrast ensures that knowledge is only available to those contributing members within the alliance; therefore, the ability for a researcher or firm outside of the alliance to pool internal knowledge with that from the closed pool may not be possible or at a cost that will vary with the market power of the closed group. All but 3 of the consortia used an open access alliance structure. The Accelrys Combinatorial Chemistry Consortium, the Accelrys Functional Proteomics Consortium, and the Accelrys Nanotechnology Consortium were all closed access consortia —ensuring that knowledge was only available to consortia members (Accelrys, 2007); (Table 2, see appendix 1).
Rules for Participation. In terms of participation, 18 consortia established rules regarding membership. Offering monetary commitments, making formal commitments to the mandate and policies of the initiative, or licensing products used within the initiative, were signals of cooperation used when joining these consortia (Table 2, see appendix 1).
While the majority of consortia allowed members with the requisite research experience to join voluntarily, 7 of these 18 initiatives used formal invitations or applications, steering or executive committees, or by-laws to determine membership. Where formal commitments were required, as for the International Regulome Consortium, participation by-laws and agreements tended to address both admission policies as well as exit policies.
Ten consortia required a monetary com- mitment as part of membership; out of this group, 2 required the maintenance of grants and 8 required up-front membership fees. In open access initiatives such as the SNP Consortium, large upfront payments were made to support research (Davies, 2001). In other instances, such as the International Structural Biology Consortium, membership fees were paid, as verified by the director in our survey. These membership fees entitled a member access to beta-version software, experimental instruments, and technology developed by associated research labs and institutions.
Both monetary fees and software licenses were required to join the Accelrys Consortia. As Accelrys software formed the basis of the consortium project, in order to take part in and obtain the benefits of the project work, members were required to maintain licenses to a number of products which formed the core of consortium technology (Accelrys, 2007).
Focus of Knowledge Production Activities. In their analysis, Allarakhia et al. (2008) determined that almost half of the 39 consortia were focused on genomic or proteomic research; an additional 7 consortia were focused on systems-based research. Interestingly, some of the consortia progressed further downstream, developing tools to support molecular biology-based drug discovery or chemistry-based drug discovery; in some cases, consortia were focused on pre-clinical and clinical research. However, only two initiatives, the Biological Innovation for Open Society (BIOS) and the Cancer Vaccine Consortium were focused on downstream biological product development (Sulston, 2006; Sabin, 2007).
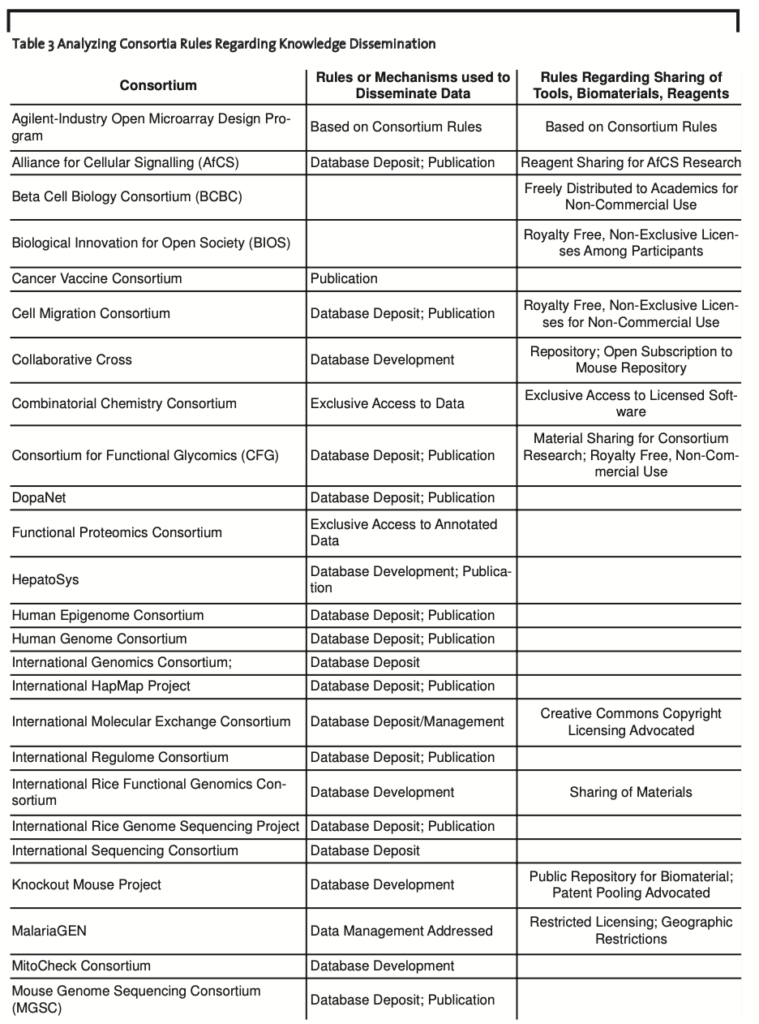
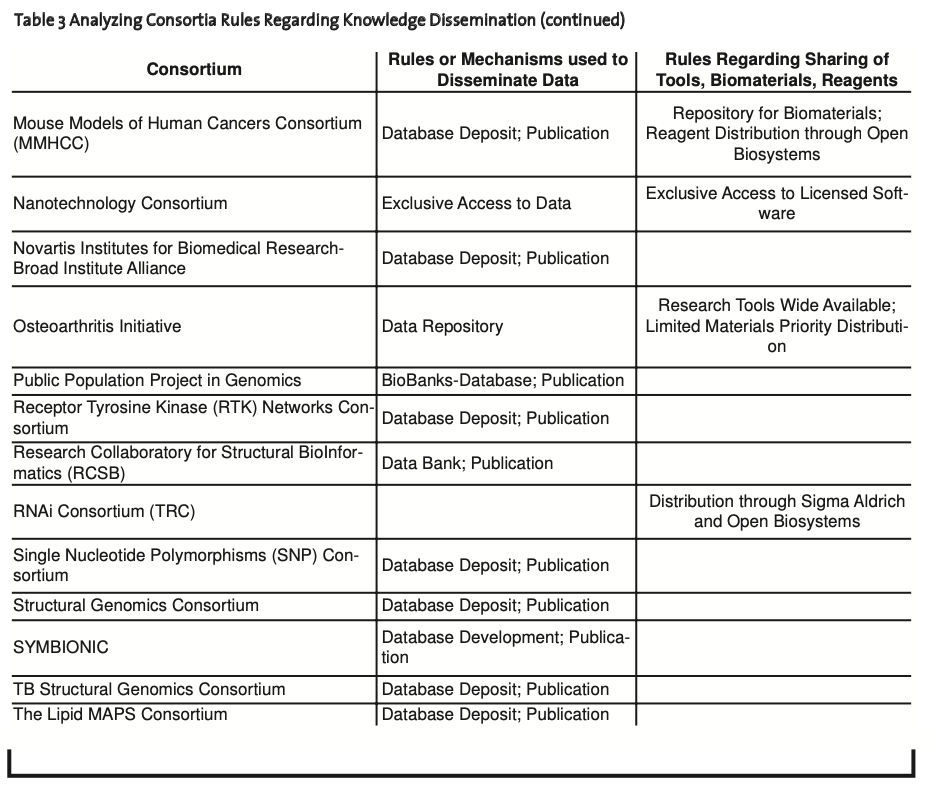
Rules for Knowledge Dissemination. In most cases, data were released almost immediately with complete access provided to members and the public at large. Data were maintained within large data repositories with the objectives of standardizing data and creating linkages between repositories developed within the consortium and between external repositories. For example, 30 consortia used or planned to use databases to provide access to upstream genomic, proteomic, systems, bio- chemical, or cell biology information. These consortia addressed the open dissemination of data as part of their rules for sharing of information with members and the public at large. In addition, 22 consortia used peer-reviewed publications to provide validated information to the public (Table 3, see appendix 2).
Allarakhia et al. (2008) were further able to determine that in the case of 16 consortia where tools, biomaterials, or reagents were either a direct outcome or a by-product of consortia member research, rules existed that addressed the sharing of these items with members or the public at large. These rules advocated sharing of materials for consortium research, ensuring access to open repositories where animal models were housed, or providing for the wide dissemination of materials for the public at large; only in a few cases was access to tools ensured for members only (Table 3, see appendix 2).
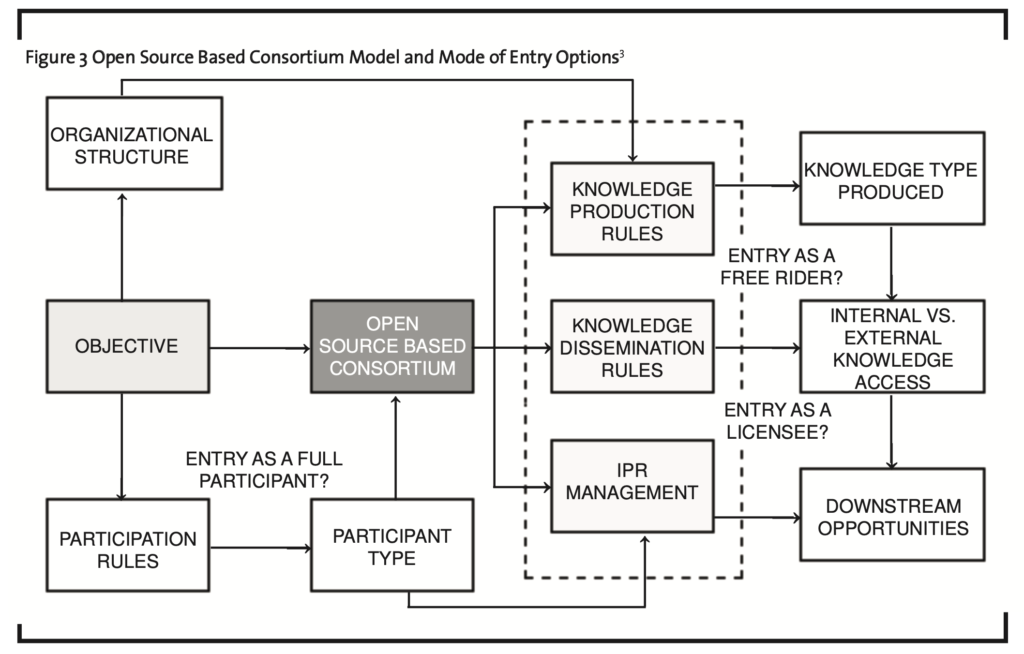
I contend that the above study provides insight for firms considering entry into the biopharmaceutical entry. Specifically, in terms of mode of entry into the biopharmaceutical arena, firms from emerging markets can enjoy many of the benefits associated with participation in open source based consortia. These benefits include early access to knowledge, absorptive capacity development, and cost sharing during knowledge production activities. However, firms from emerging markets may have limited resources available and should be aware of the structure and rules associated with the consortium before committing these resources. The objectives of the open source consortium in terms of knowledge production and dissemination will determine both the most effective organizational structure and the types of participants that will join. The organization of knowledge production activities will impact not only accessibility to knowledge but also the learning experience for any firm. An awareness of participant type — public or private sector — can enable for a determination of motivation with respect to participation and likely adherence to the open source model. Rules for participation should be understood at the outset as a monetary commitment may be required to join the consortium; in this sense, participation rules can determine which firms can join the consortium as a function of resource availability. Rules regarding knowledge access — ranging from open access for members only, to open access for members and the public at large, to open licensing, will further drive the decision to join the consortium. Depending on the knowledge access policy, a firm may be forced to join the consortium in order to access critical knowledge, a firm may choose to freeride and access knowledge without any resource commitments to the consortium, and/or choose to access knowledge as a licensee (see Figure 3).
Open Licensing as a Mode of Entry
The choice of exclusively licensing or non-exclusively licensing a patent is a function of the characteristics associated with the knowledge, the desire of the patent holder to maximize revenue from disembodied versus embodied knowledge, and the desire to diffuse the knowledge versus develop the knowledge (Arora and Fosfuri, 2003; Foray, 2004). For example, the decision to sell disembodied knowledge in the form of patents and licenses can complement or substitute for the sale of embodied knowledge. Substitution occurs when the profits from the sale of disembodied knowledge are greater than those from the sale of embodied knowledge (Antonelli, 2003; Arora and Fosfuri, 2003). Specifically, when the costs of internal coordination of the knowledge are larger than the transaction costs associated with the market for technical knowledge, or when special assets are required to progress further downstream, the patent holder may choose to maximize revenue through a licensing strategy, specifically an exclusive licensing strategy (Teece, 1986; Anto- nelli, 2003; Arora and Fosfuri, 2003).
Complementarity between the sale of disembodied knowledge and internal embodiment occurs when knowledge possesses high applicability and it is possible to operate in different markets from other licensees of the knowledge (Teece, 1986; Arora and Fosfuri, 2003, Foray, 2004; Scotchmer, 2004). In this case, a non-exclusive licensing strategy can ensure that multiple participants can pursue several streams of research. Furthermore, cross-licensing is a useful innovation management strategy when knowledge exhibits high levels of complementarity (Shapiro, 2001). With downstream activities dependent on the recombination of a variety of knowledge, the cost of coordination including accumulation of the full range of required knowledge may be too high for one innovator (Antonelli, 2003; Burk and Lemley, 2003). Namely, the capabilities of the one innovator may only cover a portion of the research domain. Consequently, innovators may find it profitable to engage in cross-licensing for knowledge. However, the ability for each innovator to access knowledge depends on the amount and type of proprietary knowledge each one is able to contribute in any bargaining event (Antonelli, 2003).
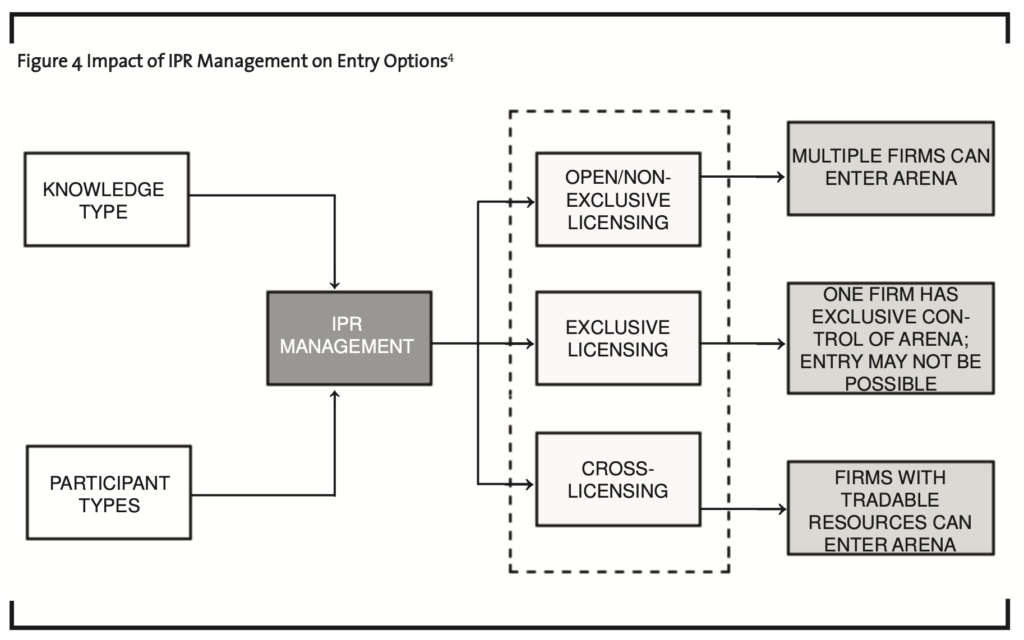
In Figure 4, I contend that both knowledge type — disembodied versus embodied — as well as participant type — private or public sector — will impact the intellectual property rights management strategy adopted. Open or non-exclusive licensing with or without royalties will encourage multiple firms to enter and/or stay within a technological arena. In contrast, exclusive licensing will enable one firm to enter and possibly maintain control of a technological arena (Walsh et al., 2003). In the case of cross-licensing, only firms with tradable knowledge assets may be able to bargain for other knowledge assets and in turn, enter or stay within the technological arena. It is important to note that the ability to enter and stay in a technological arena will also depend on the substitutability of knowledge assets. For example, the existence of non-infringing workaround solutions will encourage a licensor to provide non-exclusive licenses (Allarakhia et al., 2008; Antonelli, 2003).
In their study of biopharmaceutical consortia, Allarakhia et al. (2008) found that consortia differentiated between disembodied knowledge in the form of raw data and embodied knowledge created by consortia members in the form of tools, biomaterials, and reagents. Although disembodied data was mandated in most cases for almost immediate release, tools, biomaterials, and reagents could be appropriated and licensed to consortia members and the public at large. Appropriation activities were regulated by the provision of rules regarding licensing terms. Supporting data and materials sharing policies provided by the NIH, the Wellcome Trust, the Creative Commons, the Biological Innovation for Open Society, and even private sector firms such as Open Biosystems, enabled for relatively easy access to disembodied and embodied knowledge created within consortia (Table 3, see appendix 2).
From the consortia analysis, Allarakhia et al. (2008) were able to identify various licensing agreements employed to widely disseminate embodied knowledge as well as copy-righted material (Table 4). In each instance, the objective was to ensure that multiple firms would have the incentive to enter and remain within the technological arena.
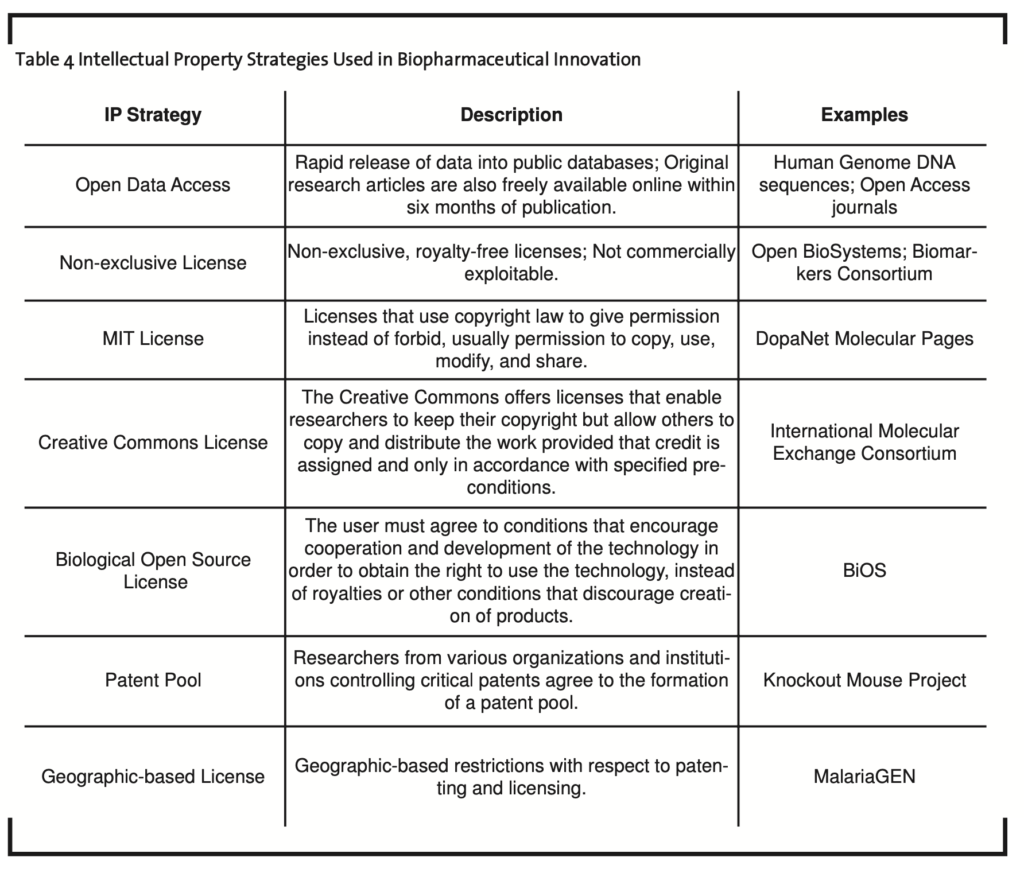
Non-exclusive license. Many instances were found of non-exclusive, royalty-free licenses used to disseminate knowledge generated by consortia members. In one instance, a limited use license provided researchers with a limited, non-exclusive, non-transferable right to the product (with no right to resell, repackage, or further sublicense). For example, the purchase of products distributed through this licensing agreement did neither include nor carry any right or license to use, develop, or otherwise exploit products commercially (Open Biosystems, 2007). In the case of one consortium, members could offer royalty-free licenses for tools and data to project team members, and royalty-free licenses for non-commercial use to others (Biomarkers Consortium, 2008).
MIT license. DopaNet’s Molecular Pages is a collection of annotated quantitative biotechnology data. DopaNet Molecular Pages are available under the terms derived from the MIT License (Le Novère and Donizelli, 2004). The MIT License, also called the X License or the X11 License, originated at the Massachusetts Institute of Technology, and is a license for the use of certain types of computer software; essentially, a non-copyleft (licenses that use copyright law to give permission instead of forbid) free software license. The license allows a user to deal with the software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the software (Open Source Initiative, 2007).
Creative Commons license. The International Molecular Exchange Consortium applies the Creative Commons Attribution License. This consortium is a group of major public interaction data providers sharing curation efforts and exchanging completed records on molecular interaction data. The Creative Commons offers licenses that enable researchers to keep their copyright but allow others to copy and distribute the work provided that credit is assigned and only in accordance with specified pre-conditions including attribution plus non-commercial use only, attribution and non-derivative use, or attribution and dissemination to others that follow the same conditions set by the original researcher (Creative Commons, 2007; International Molecular Exchange Consortium, 2007).
CAMBIA-Biological Open Source. The Biological Innovation for Open Society (BiOS) is an initiative of the Center for Applications of Molecular Biology in Agriculture (CAMBIA) with the objectives to develop new means for cooperative invention, improvement, and delivery of technologies for life sciences. The licensing strategy promoted by BiOS hopes to encourage entry into a technological arena with a focus on those researchers and firms in neglected markets. Specifically, it is anticipated that open source agricultural research will enable innovation by small biotechnology companies. This will enable the development of locally suited technologies, reduce dependence on giant agribusiness conglomerates, and facilitate research on crops suited for local conditions in the developing countries (Thomas, 2005).
Under a BiOS-compliant agreement, the user must agree to conditions that encourage cooperation and development of the techno- logy in order to obtain the right to use the technology, instead of royalties or other conditions that discourage creation of products (Sulston, 2006). The conditions include a provision that licensees cannot exclusively appropriate the fundamental essence of the technology or improvements (BiOS Initiative, 2007). The base technology remains the property of the entity that developed it, but improvements can be shared with others that support the development of a protected commons around the technology; participants who agree to the same terms obtain access to improvements and other information, such as regulatory and biosafety data (BiOS Initiative, 2007). To maintain legal access to the technology, users must agree not to prevent others who have agreed to the same terms from using the technology and any improvements in the development of varied products.
Patent pool. It is anticipated that the Knock- out Mouse Project will require the resolution of several intellectual property claims involving both the production and use of knockout mice. The Knockout Mouse Project is an initiative that aims to generate a comprehensive and public resource comprised of mouse embryonic stem (ES) cells containing single deletions (knockouts) of every gene in the mouse genome — essentially research tools to understand the role of genes in biological processes. Hence, researchers from various organizations and institutions controlling such patents have agreed to the formation of a patent pool of mouse knockout technologies to enable the development of these stem cells (Austin et al., 2004).
Geographic-based licensing. The Grand Challenges in Global Health, which funds MalariaGEN, has developed the Global Access Strategy. This system requires grantees to prepare both a strategy for commercialization of research and an intellectual property management policy. Key provisions of the Global Access Strategy include a requirement that the principles of the strategy apply to licenses and con- tracts that use intellectual property of the consortium; that downstream licensees of the consortium’s intellectual property not apply for secondary patents in the developing world that would prevent access to affordable health care solutions; and a stipulation that prohibits exclusive licensing of the consortium’s intellectual property except in cases where it is necessary to provide a marketing incentive (Chokshi et al., 2006).
Discussion
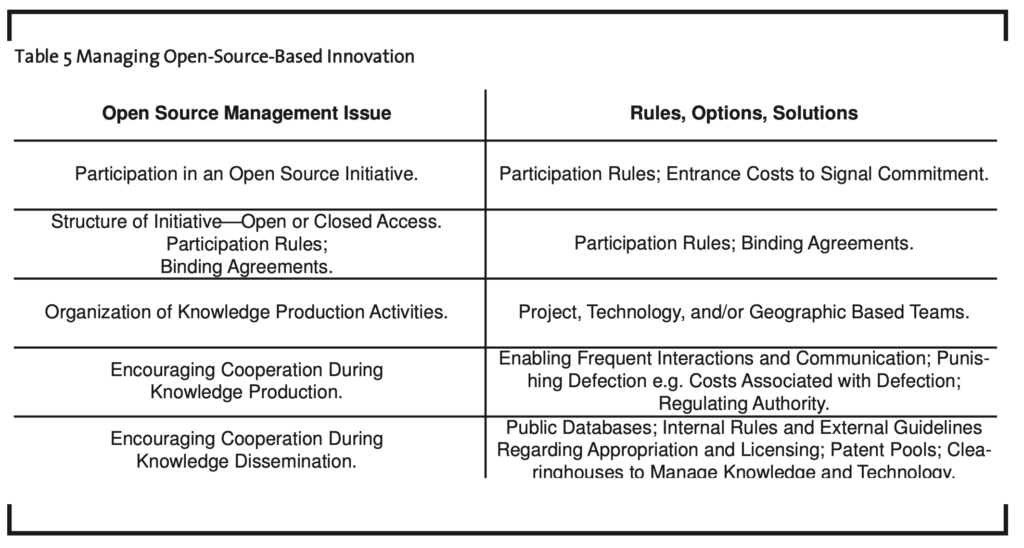
The ability to join an open source initiati- ve will be tempered by informal and formal rules of participation. With formality, entrance costs may be used to facilitate research and development activities as well as to signal cooperation and commitment to the initiative (Kollock, 1998; Gintis et al., 2001). The role of such entrance costs or rules for participation is to create trust through a visible signal. For example, committing resources in advance including monetary fees makes other participants in the initiative, and future researchers who are considering participation, aware of a researcher’s cooperative intentions (Gulati et al., 1994). The decision to participate in any initiative is also affected by the degree of accessibility to the associated knowledge. Open access ensures that knowledge will be available to all participants in future downstream research regardless of participation (Gintis et al., 2001). Closed access in contrast, ensures that knowledge is available only to contributing members within the initiative.
In terms of property rights, Ostrom argues these rights do not emerge spontaneously from a common property system. Private property rights depend on the existence and enforcement of rules that define who has a right to pursue which activities involving a resource and how the returns from that activity will be allocated (Ostrom, 1989). For example, the use of binding agreements can ensure cooperation during knowledge dissemination. Therefore, in the management of open source initiatives, the research outcomes to be disseminated, the format for dissemination, and the knowledge to be privatized, should be clearly understood by all the participants. Internal rules or mechanisms used to promote cooperative behaviour can include: formalizing the requirements to join the knowledge network, ensuring frequent interactions, encouraging communication between participants, punishing defection, and setting the boundary for access to knowledge. An authority that regulates access to knowledge can ensure that a fair and efficient knowledge governance strategy is indeed used.
If and when knowledge is appropriated through the filing of patents, rules should further encourage licensing that provides the greatest collective value to the initiative members and/or the public at large. For example, many of the consortia analyzed by Allarakhia et al. (2008) advocated the use of royalty-free non-exclusive licenses. Where technology can be substituted through non-infringing workaround solutions, a patent holder will also have an incentive to offer a non-exclusive license, rather than face competition without any possible compensation for his/her initial discovery. Alternatively, in cases where the market for technology is relatively small with technology having zero standalone commercial value, a patent holder may need to offer a non- exclusive license to ensure that a downstream developer will use the technology in products, thereby enabling the patent holder to reap the rewards of his/her original discovery.
From a mode of entry perspective, open source initiatives can level the playing field for new entrants into a technological arena. Organizations from emerging markets adhering to the open source model should equally ensure that the public domain of knowledge is not only sustained, but also augmented.
Public-sector and private sector organizations from such economies can institute policies that preserve the public domain of knowledge, enable the formation of open source initiatives for standard or technology development, encourage the use of open licensing strategies for appropriated knowledge, and the use of clearinghouses that can manage knowledge and technological assets — ensuring broad dissemination and adoption of these assets. Table 5 outlines these issues and the rules or associated solutions that can be used to manage open-source-based initiatives.
Conclusion
Rising costs, technological complexities, and shorter life cycles have put pressure on companies and their internal innovation processes. Chesbrough (2003) discusses that open business models can enable biopharmaceutical companies to leverage external resources and human capital to save time and money during the innovation process. The open business model further allows companies to generate revenue through the licensing of technologies that cannot be fully exploited within an organization and through the in-licensing of technologies that are discovered outside the boundaries of the organization (Chesbrough, 2003). Therefore, managers of firms in deve- loped and in emerging markets alike should seek out these opportunities presented by open innovation — including participating in open source based innovation.
For firms in emerging markets, open source based innovation presents a cost-effective means to learn about a domain and the corresponding product development opportunities. These firms can then use the experience gained from participation in open-source based innovation to make an informed decision regarding the investment into product development. In the biopharmaceutical industry, as product development includes expensive clinical trial testing and regulatory approvals, an informed decision needs to be based on the firm’s resource availability across the biopharmaceutical value chain as a function of a particular technological opportunity — perhaps even the need to continue participating in open innovation during product development.
The practical lessons learned from this paper, however, indicate that firms from emerging markets with limited resources will have to carefully evaluate the objectives of an open-innovation- (including open source) based community and/or network. The objectives can include the creation of pure knowledge or even embodied knowledge in the form of tools and products. Ultimately then, firms hoping to enter a biopharmaceutical arena will have to analyze where they are located on the learning curve and what they hope to gain through participation in an open-innovation-based community. Organizational structures will then determine how firms can participate in any learning and knowledge development processes. Specifically, the distance from knowledge development activities and any supporting organizational structures that seek to minimize this distance, will determine how much learning by doing and using firms will experience. This learning by doing and using will be of particular relevance to firms from emerging markets. Finally, the mechanisms used to disclose and share knowledge will impact whether firms can indeed move down the bio-pharmaceutical value chain. It is therefore anticipated that open-innovation-based communities with clear rules, leadership, and transparent processes will be more productive — avoiding any surprises for firms with limited resources contemplating participation.
In terms of future research, it is essential to analyze new case studies involving emerging market firms and their participation in open innovation communities. These case studies should seek to look at the evolving models of open innovation as the number and type of participants change, as the objectives with respect to innovation evolve, and as the complexities associated with knowledge structures increase so that knowledge management becomes paramount. This analysis should further seek to understand any geographic-based issues hampering technological innovation by firms in emerging markets and how to eventually position these firms to meet both glo- bal and local product needs through open innovation.
References
Accelrys Nanotechnology Consortium (2007): www.accelrys.com, accessed 6/2007.
Agilent-Industry Open Microarray Design Program (2007): www.agilent.com, accessed 6/2007.
Allarakhia, M., Kilgour, D. M., Fuller D. (2008): Modeling the Incentive to Participate in Open Source Innovation, The R&D Management Conference, Ottawa, Canada, June 17-20, 2008.
Antonelli, C. (2003): Knowledge complementarity and fun- geability: Implications for regional strategy, Regional Studies, 37 (6-7), pp. 595-606.
Arora, A., Fosfuri, A. (2003): Licensing the market for techno- logy, Journal of Economic Behavior & Organization, 52 (2), pp. 277-295.
Austin, C.P. et al. (2004): The Knockout Mouse Project, Nature Genetics, 36 (9), pp. 921-924.
Bergstrom. L. (2006): Open source and open medicine take centre stage at UN research symposium, EurekAlert, accessed November 2008.
Biomarkers Consortium(2008): www.biomarkersconsortium.org, accessed 11/2008.
BiOS (2007): www.bios.net, accessed 6/2007.
Blumenthal, D., Causino, N., Campbell, E. G. (1997): Academic- industry research relationships in genetics: A field apart, Nature Genetics, 16 (1), pp. 104-108.
Bower, J. D., Whittaker, E. (1992): Global R&D networks: The case of the pharmaceutical industry, Journal of Industry Studies, 1 (1), pp. 50-63.
Burk, D. L., Lemley M. A. (2003): Biotechnology’s Uncertainty Principle, in: Perspectives on Properties of the Human Genome Project, Elsevier Academic Press, San Diego CA, pp. 305-354.
Cassier, M. (2002): Private property, collective property, and public property in the age of genomics, International Social Science Journal, 54 (1), pp. 83-98.
Chesbrough,H.W.(2007):WhyCompaniesShouldHaveOpenBusiness Models, MIT Sloan Management Review, 48 (2), pp. 22-28.
Chesbrough, H. W. (2003): The Era of Open Innovation, MIT Sloan Management Review, 44 (3), pp. 35-41.
Chesbrough, H. W., Vanhaverbeke, W., West J. (2006): Open Innovation: Researching a New Paradigm, Oxford University Press, Oxford.
Chokshi, D. A., Parker, M., Kwiatkowski, D. P. (2006): Data sharing and intellectual property in a genomics epidemio- logy network: Policies for large-scale research collabo- ration, Bulletin of the World Health Organization, 84 (5), pp. 382-387.
Cohen, W.M., Levinthal, D.A. (1990): Absorptive Capacity: A New Perspective on Learning and Innovation, Adminis- trative Science Quarterly, 35, pp. 128-152.
Creative Commons (2007): creativecommons.org, accessed 6/2007.
Davies, K. (2001): Cracking the Genome: Inside the Race to Unlock the Human DNA, The Free Press, New York, NY. Dutfield, G. (2003): Intellectual property rights and the life sciences industries: A twentieth century history, Ashgate Publishing Limited, Burlington VT.
Foray, D. (2004): The Economics of Knowledge, MIT Press, Cambridge, MA.
Gintis, H., Alden Smith, E., Bowles, S. (2001): Costly Signaling and Cooperation, Journal of Theoretical Biology, 213, pp. 103-119.
Grant, R. M., Baden-Fuller, C. (2004): A knowledge accessing theory of strategic alliances, Journal of Management Studies, 41 (1), pp. 61-84.
Gulati, R., Khanna, T., Nohria, N. (1994): Unilateral commit- ments and the importance of process in alliances, MIT Sloan Management Review, 35 (3), pp. 61-69.
Hood, L. E. (2000): The university office of technology transfer: The inventor/researcher’s view, CASRIP Symposium Publication Series, No. 5, CASRIP, University of
Washington, Seattle WA.
International Molecular Exchange Consortium (2007): imex.sourceforge.net, accessed 6/2007.
Kitano, H. (2002): Systems biology: A brief overview, Science, 295 (5560), pp. 1662-1664.
Kitano, H. (2001): Systems biology: Toward systems-level understanding of biological systems, in: Kitano, H. (ed.), Foundation of systems biology, MIT Press, Cambridge, MA, pp. 1-29.
Kollock, P. (1998): Social dilemmas: The anatomy of coopera- tion, Annual Review of Sociology, 24, pp. 183-214.
Lakhani, K. R., von Hippel, E. (2003): How Open Source Soft- ware Works: Free User-to-User Assistance, Research Policy, 32 (6), pp. 923-943.
Lawler, A. (2004): Broad-Novartis Venture Promises a No- Strings, Public Gene Database, Science, 306, p. 795.
Le Novère N., Donizelli M. (2004): The Molecular Pages of the Mesotelencephalic Dopamine Consortium (DopaNet), BMC Bioinformatics, 5, pp. 174.
Lemley, M. (2002): Intellectual Property Rights and Standard-Setting Organizations, California Law Review, 90, pp. 1889-1981.
Malhotra, N. (2003): The Nature of Knowledge and the Entry Mode Decision, Organization Science, 24 (6), pp. 935-959.
Marshall, E. (1997): The battle Over BRAC1 goes to court, BRAC2 may be next, Science, 278 (5345), pp. 1874.
Meredith, L. (2005): BRIC by Brick; IBM looks to open source in emerging markets, SearchDataCenter, February 24
Appendix